Barcoding of the Fungarium YSU collection
The Molecular Laboratory enabled the start of DNA barcoding of the Fungarium. The laboratory is equipped with several zones, including extraction, PCR, electrophoresis, purification and Sanger sequencing zones. The laboratory is staffed by qualified technicians and researchers.
The YSU Fungarium collection was created as part of the revision of the diversity of fleshy fungi of West Siberian middle taiga and contains about 10 thousand dried specimens. Special attention has been paid to the communities of raised bogs, which are widespread in the region. The barcoding roject is designed to run for three years and will include a revision of all taxonomic groups represented in the Fungarium using modern molecular genetic methods.
Main stages of collection barcoding:
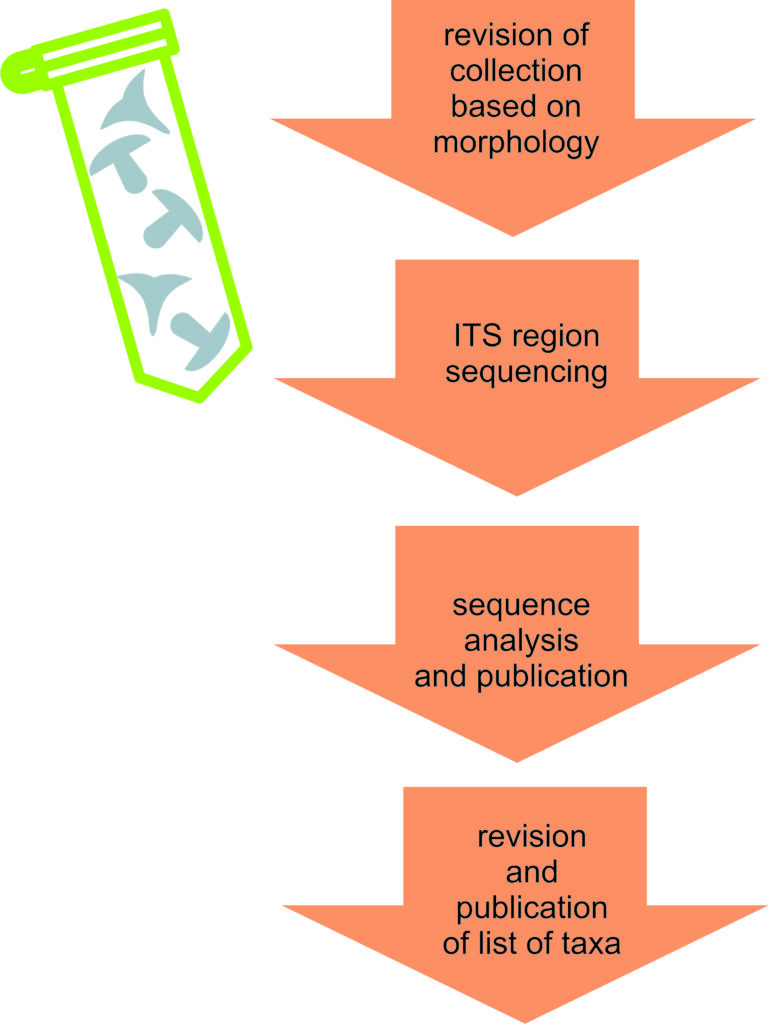
- Morphological revision of collections of a specific group, uploading of descriptions and photographs into the YSU Fungarium database;
- Selection of 2-3 specimens of each morphological taxon for subsequent sequencing;
- All stages of processing from extraction to sequencing with working data recorded in the Laboratory Journal database;
- Quality assessment, closest match analysis and saving of sequence data in the Fungarium database;
- Publishing of sequences in open data portals (GenBank, GBIF, PlutoF);
- Analysis of taxonomic group and publication as checklists, new finds or monographs.
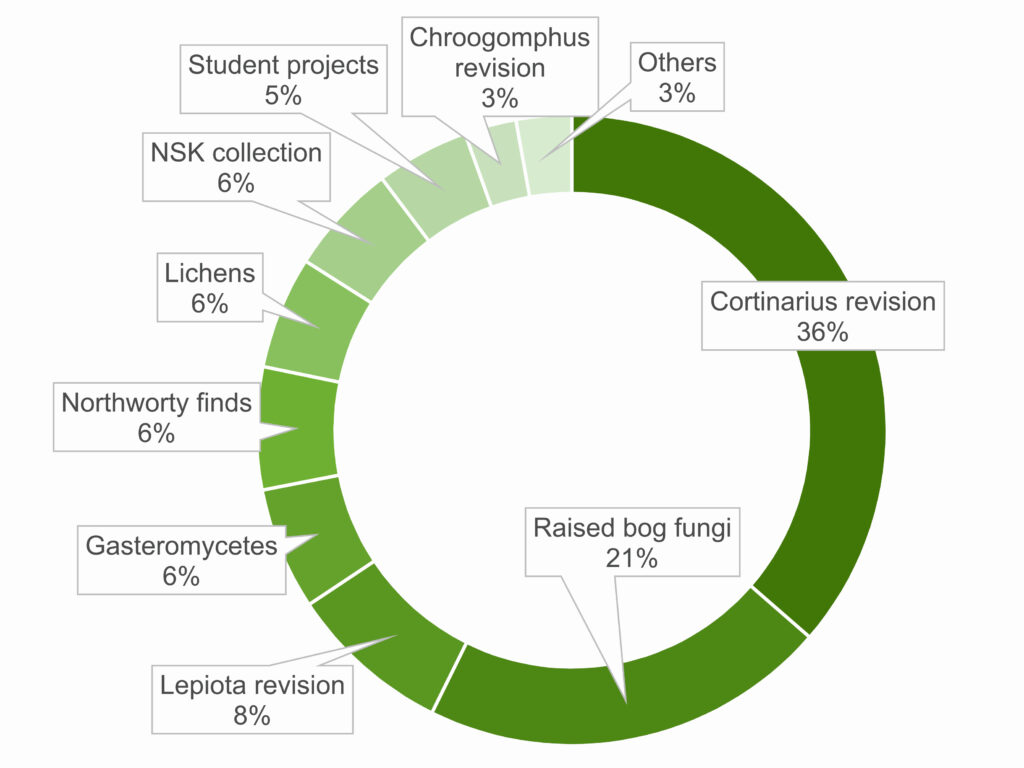
During the two years of the project, over 1500 extractions from about 1000 specimens were performed. About a third was a part of the revision of the large genus Cortinarius. Another large part was within the effort of raised bog fungi barcoding. The remaining specimens were from revisions of minor groups, students projects and sequencing of noteworthy finds and taxa potentially new to region or to science in general.
Storage of sequences in databases
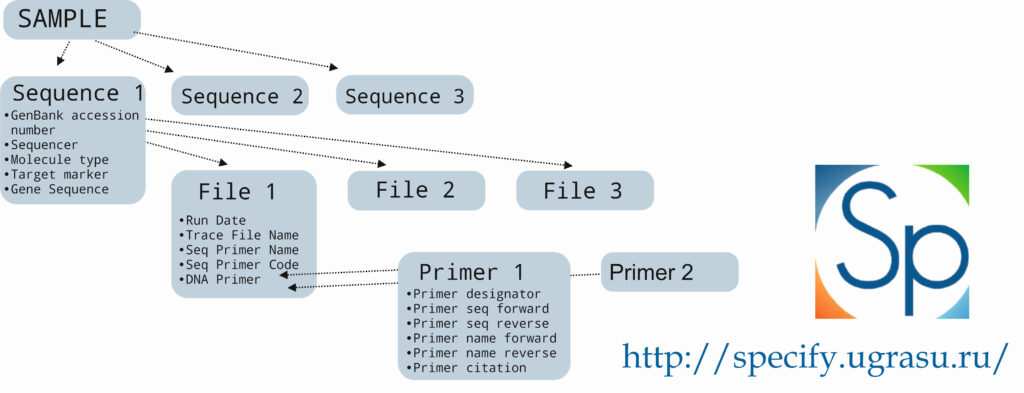
Ready sequences are uploaded to the YSU Fungarium database in Specify 7. Sequence metadata, the edited sequence and original files are saved. Each sample may correspond to one or several sequences (different regions). Each sequence may have one or several original files with their sequencing parameters. Primer parameters are filled in a separate table.
Sequences are published on the following websites
- GBIF (YSU Fungarium dataset)
- GenBank
- PlutoF
- BOLD
Published datasets
- Filippova N (2023). The Fungarium of Yugra State University. Version 1.128. Yugra State University Biological Collection (YSU BC). Occurrence dataset https://doi.org/10.15468/g4bk6h accessed via GBIF.org on 2023-08-16.
- Filippova N, Rudykina E, Zvyagina E (2023). The checklist of macrofungi of raised bogs: barcoding of accumulated collection following the 9-year plot-based monitoring in Northwestern Siberia. Version 1.3. Yugra State University Biological Collection (YSU BC). Occurrence dataset https://doi.org/10.15468/dl.gebkc9 accessed via GBIF.org on 2023-08-15.
- Filippova N, Zvyagina E, Rudykina E (2023). BOLD dataset: Barcoding of Macromycetes from Raised bogs in Northwestern Siberia. https://doi.org/10.5883/DS-BFSEG
Published papers
- Filippova N, Zvyagina E, Rudykina EA, Ishmanov TF, Filippov IV, Bulyonkova TM, Dobrynina AS (2024) DNA-based occurrence dataset on peatland fungal communities studied by metabarcoding in north-western Siberia. Biodiversity Data Journal 12: e119851. https://doi.org/10.3897/BDJ.12.e119851
- Kotkova V. M., Afonina O. M., Alverdiyeva S. M., Anissimova O. V., Bragin A. V., Cherenkova N. N., Davydov E. A., Dongak D. A.-S., Doroshina G. Ya., Efremov A. N., Filippova N. V., Gorbunova I. A., Himelbrant D. E., Kapitonov V. I., Korchikov E. S., Kurbatova L. E., Kuzmina E. Yu., Makarova O. L., Mongush Ch. B., Moroz E. L., Moseev D. S., Notov A. A., Novozhilov Yu. K., Plikina N. V., Popova N. N., Romanov R. E., Safronova T. V., Shadrina S. N., Shiryaeva O. S., Stepanchikova I. S., Storozhenko Yu. V., Tarasova V. N., Tsurykau A. G., Vaishlya O. B., Vishnyakov V. S., Vlasenko A. V., Vlasenko V. A., Yakovchenko L. S., Yarutich I. A., Zhamangara A. K., Zhuykov K. A., 2024. New cryptogamic records. 13. Novosti sistematiki nizshikh rastenii 58 (1): R1-R45. https://doi.org/10.31111/nsnr/2024.58.1.R1
- Filippova N. V., Gorbunova I. A., 2024. Two species of Amanita (Basidiomycota) new to Russia from the South Siberia. Novosti sistematiki nizshikh rastenii 58 (1): F37-F46. https://doi.org/10.31111/nsnr/2024.58.1.F37
- Ageev DV, Bulyonkova TM, Zvyagina EA, Filippova NV, 2024. Hohenbuehelia incrustata sp. nov., a new species of Hohenbuehelia (Pleurotaceae, Agaricales) from Northwestern Siberia, Russia // Phytotaxa [submitted to the journal]
- Zvyagina EA, Sazanova NA, 2024. Suillus kovalenkoi (Boletales, Agaricomycetes), a new Larix-associated species from North Asia // Phytotaxa [submitted to the journal]
- Filippova N.V., Zvyagina E.A., Bolshakov S.Yu., Rudykina E.A., Dobrynina A.S., 2023. The diversity of larger fungi of peatlands: nine years of plot-based monitoring with barcoding of revealed species in a raised bog “Mukhrino”, West Siberia // Biodiversity Data Journal, 11: e105111, https://doi.org/10.3897/BDJ.11.e105111
- Kotkova V. M., et al., 2023. New cryptogamic records. 12 // Novosti sistematiki nizshikh rastenii, 57(2), https://doi.org/10.18822/edgcc472141
