Metabarcoding of fungal communities in taiga zone of Western Siberia
A program on metabarcoding of fungal communities in different types of ecosystems in the taiga zone of Western Siberia was initiated. The first part of the project focuses on metabarcoding various substrates in raised peatand ecosystems. Currently, we are expanding the diversity of habitats by sampling soil and litter from different types of forests, studying fungal communities in phylloplanes, and investigating changes in species composition under the influence of oil spills.
Two different approaches to eDNA sequencig are used: Illumina MiSeq (based on outsourcing DNA-samples) and Nanopore MinION technologies.
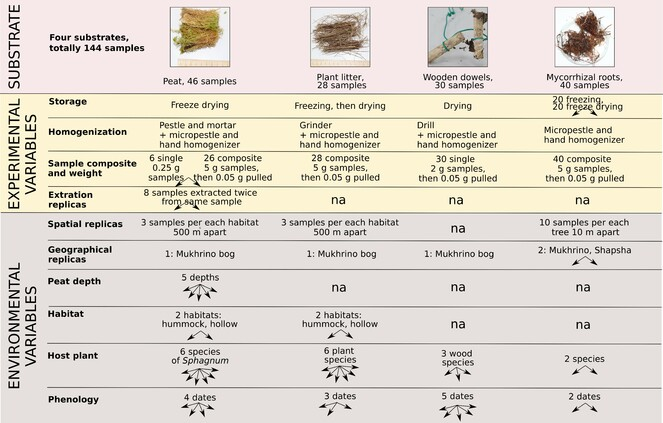
To supplement long-term monitoring of fungal communities of the raised peatland with an environmental DNA approach, we completed a series of samplings of common substrates in the same locality in the Mukhrino Bog. Four major substrates were subjected to metabarcoding analysis: peat (from the surface layer to a depth of about 3 m), leaf litter of six bog plants, wood (represented by standardised wooden dowels) and mycorrhizal roots of two bog-dwelling trees.
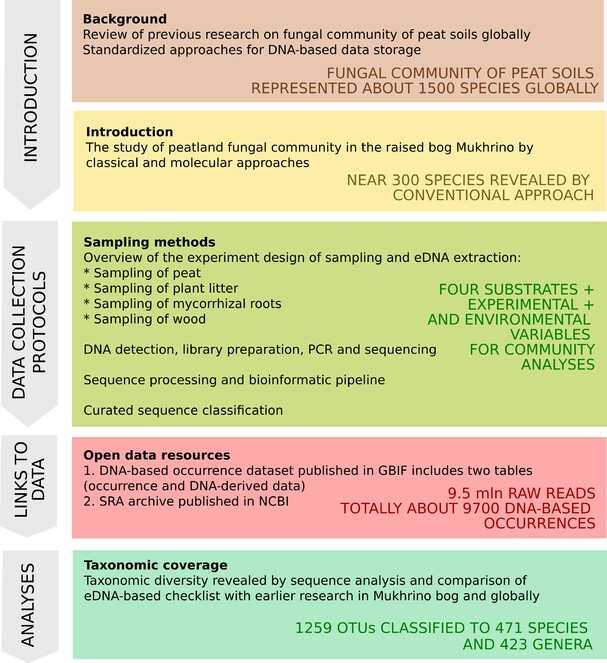
Metabarcoding of the ITS2 region (Illumina MiSeq platform) revealed about 1200 OTUs and 800 Linnean taxa. The obtained sequences were processed using QIIME2 (Quantitative Insights Into Microbial Ecology 2, version 2023.9).
The sequence analysis revealed a total of 1259 OTUs classified into 471 species, 423 genera, 223 families, 86 orders, 30 classes, seven phyla and one kingdom at a 99% similarity level. About 42% of taxa were identified at the species level, 21% at the genus level and the rest at higher taxonomic levels.
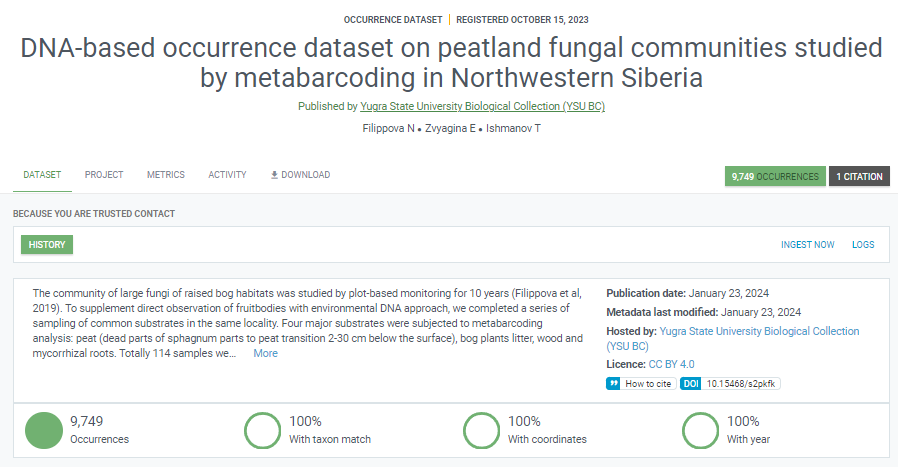
The dataset representing DNA-based occurrences published in GBIF as an occurrence dataset with the DNA-derived extension table based on guidelines (Filippova et al. 2023b).
Last update: 09/04/2024
